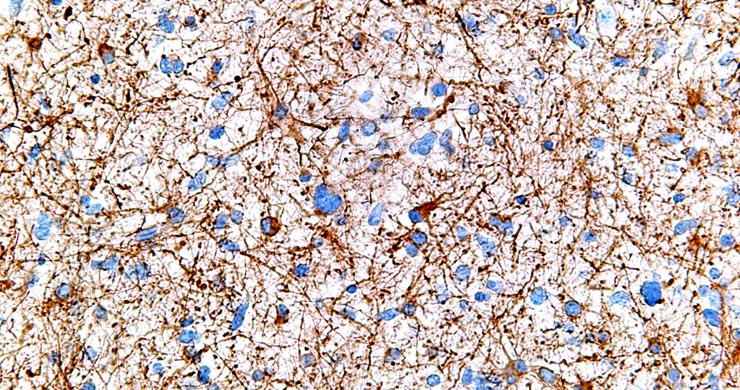
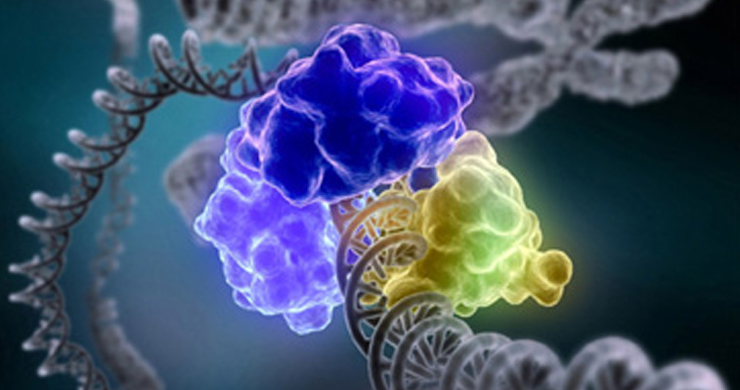
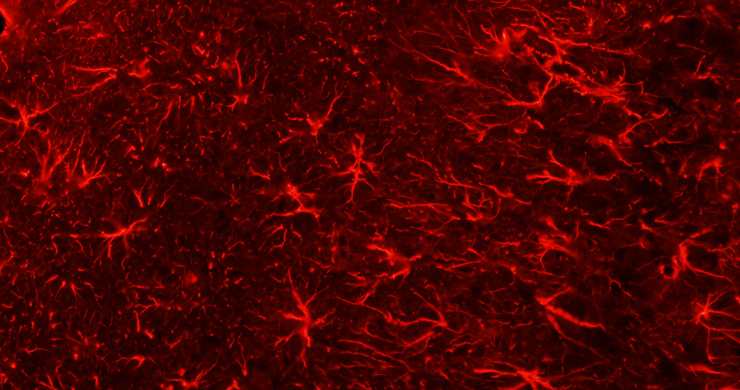
Our research aims to understand how readout of the information encoded in the DNA of plant and animal genomes can be influenced by the epigenome, the extent to which epigenomic modifications such as DNA methylation can be altered by the environment and in disease states, and to develop molecular tools for precision epigenome engineering.
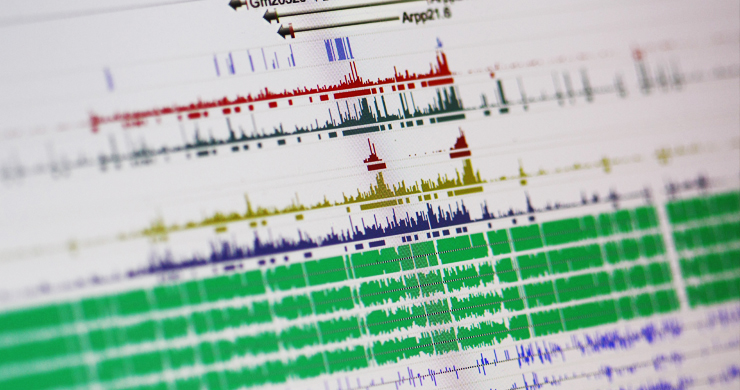
By modulating accessibility to the information encoded in the genome, chemical modifications of the DNA and associated proteins can affect gene activation and repression to execute distinct transcriptional programs and impart a stable state of transcriptional activity. We use advanced DNA sequencing, molecular, genetic and computational techniques to generate whole-genome high resolution maps of epigenetic modifications in a diverse range of complex multicellular organisms, including plants, humans, mice, and diverse vertebrates. Through these studies we aim to elucidate the mechanistic underpinnings of how the epigenome is established and dynamically modified, and how it affects the cellular readout of the underlying genetic information.
Developing a comprehensive understanding of how the cell utilizes the epigenome is essential in order to both understand the roles it plays in eukaryotic development and stress response, and to develop effective strategies to remedy its disruption in disease states.
Our research is generously supported by funding from the Australian Research Council, the National Health and Medical Research Council, HHMI, The Sylvia and Charles Viertel Charitable Foundation, and The University of Western Australia.